Genomic Solutions for Your Solid Tumor Project
At Cerba Research, our expert team can optimize solid tumor trials due to our extensive experience in genomics and access to a full range of sophisticated instruments and robust broad panel assays. We are also continuously verifying solid tumor international guidelines and adapting our existing panels accordingly.
Our experienced scientific team provides a flexible and responsive approach to your solid tumor trial. Note that about 50% of our oncology trials are solid tumor driven currently and may require broad panel NGS assays.
Need to assess the total number of somatic mutations? Or determine the alteration of one or multiple nucleotides of your DNA sequence? Thanks to Cerba Research and Cerba Healthcare’s experience and history in genomics, we are able to propose broad panel NGS assays. These may cover the majority of gene alterations with established, emerging, and exploratory value across lung, ovarian, breast, colon, melanoma, bladder, GIST, and rare tumors (sarcomas).
Additionally, those panels may also determine HRD score, TMB status, and MSI. Panels may detect many hallmarks of cancer such as, but not limited to:
- Single nucleotide variants (SNVs)
- Insertions-deletions (indels)
- Copy number variations (CNVs)
- Gene fusions (translocations)
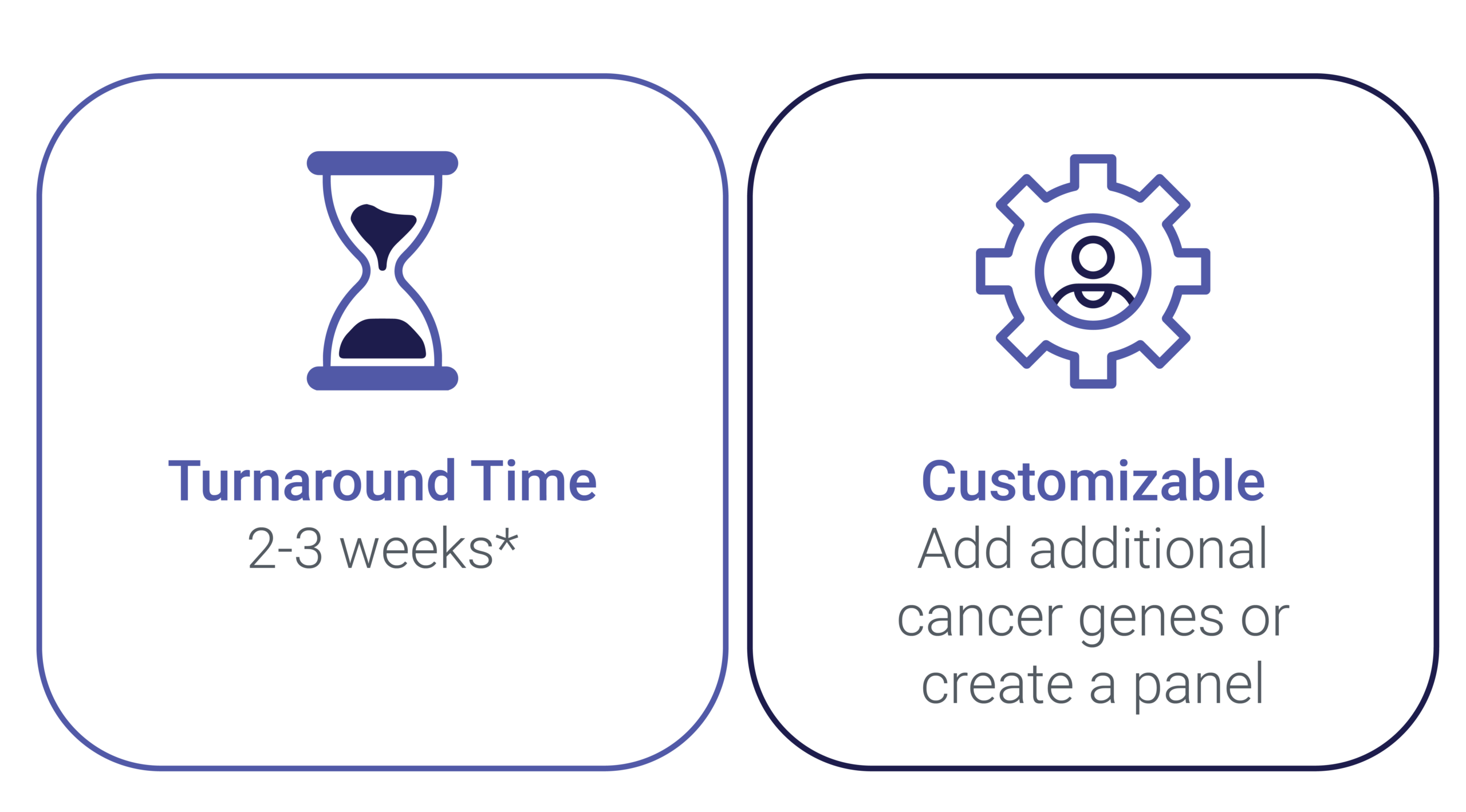
Customize a New Panel According to Your Trial Needs
Allow your scientific project a sufficient amount of time for analytical validation. If an NGS assay is already in accordance with your trial needs, the turnaround time from receipt of approved samples to result is normally 10-15 days.
Our NGS For Solid Tumors
A full range of capabilities and an Scientific team dedicated to your trial
Cerba OncoSign Panel
42 DNA
17 RNA (fusions)
15 microsatellites
Hallmarks of cancer
Cerba OncoSign panel covers mutations with established and emerging value across lung, ovarian, breast, colon, melanoma, bladder, GIST and more. It may also determine MSI-H and may be performed on FFPE (OncoSign) & liquid biopsy (OncoSign ctDNA). It is also performed on FFPE for routine clinical use in Europe.
EGFR (FFPE / liquid biopsy)
Endopredict
BRAF V600E
Amplifications: MET / HER2
Fusions: ALK / ROS1 / RET / NTRK1-2-3
Microsatellite instability (MSI) status may be provided by using the OncoSign panel.
Molecular Analysis in Non-small Cell Lung Cancer (NSCLC)
With precision medicine becoming a more integral component of NSCLC treatment, molecular testing is an important step to help researchers, pathologists, and oncologists understand the genetic underpinnings of this devastating disease.
Discover how Cerba Research envisions the usage of an NGS-based broad panel assay to detect clinically relevant genetic alterations in NSCLC. Read our article on Precision medicine for NSCLC to find out more.

Cerba OncoSign 600+ For Solid Tumors
Benefit from Cerba OncoSign 600+, our extra large NGS panel which may detect 638 DNA-based genes including 20 fusion genes. Cerba OncoSign 600+ panel covers mutations with established, emerging and exploratory value across,lung, ovarian, breast, colon, melanoma, bladder, GIST, rare tumors and more It will also determine MSI & TMB (HRD coming soon).
Microsatellite instability (MSI) is a modification that occurs in cancer cells where the number of repeated DNA bases in a microsatellite (a short sequence of DNA) is different from what it was when the microsatellite was inherited. MSI may be caused by DNA mistakes that are not corrected and is found most often in colorectal, cancer, and endometrial cancer. Knowing whether a cancer has high MSI may be a predictive marker for response to checkpoint inhibitor regimens or clinical trial options.
Tumor Mutational Burden (TMB) is an approximate measure of the total number of somatic mutations. Theoretically, high TMB levels will correlate with high neoantigen levels that will activate an antitumor immune response. TMB is currently an emerging immune biomarker for patients with metastatic non-small cell lung cancer based on clinical data. For example, tumors that have a high TMB appear to be more likely to respond to certain types of immunotherapy
A fusion gene is the juxtaposition of two otherwise separate genes, the transcription of which is translated into an abnormal protein (often a fusion protein that leads to a constitutive protein tyrosine kinase) that drives the growth of many cancers. The detection of fusion genes, also known as gene rearrangements, are important in targeted therapy clinical development and a great example of precision medicine. Such fusion genes may be ALK, ROS1 and NTRK1-3 to name a few.
Copy number variants (CNVs) refer to a circumstance to which the number of copies of a specific segment of DNA varies among different individuals’ genomes. Genetic variants, including insertions, deletions, and duplications of sections of DNA, are collectively referred to as CNVs. CNVs account for a large proportion of genetic variation between individuals and present a unique opportunity to investigate the impact of CNVs in drug development.
Single nucleotide variants (SNVs) is a DNA sequence variation that occurs when a single nucleotide, such as adenine, thymine, cytosine, or guanine, is altered. SNVs are sometimes referred to as single nucleotide polymorphisms.
The study of SNVs in clinical trials is relevant in providing information on disease risk assessment.
Cerba Paris: Our Comprehensive Oncopanels
| Genes | Cancer Type | Instrument | TAT* | Bone marrow | ctDNA | FFPE | |
|---|---|---|---|---|---|---|---|
|
Cerba OncoSign |
59 |
Solid tumors (important hallmarks of cancer) 42 DNA 17 RNA (fusions) 15 microsatellites HRD status (CE-IVD marked & in a separate panel) |
NextSeq |
15 |
X |
X |
|
|
Cerba OncoSign 600+ |
658 |
Solid tumors (important hallmarks of cancer) 638 DNA 20 RNA (fusions) TMB, MSI, HRD |
NextSeq |
15 |
X |
||
| Genes |
59 |
|---|---|
| Cancer Type |
Solid tumors (important hallmarks of cancer) 42 DNA 17 RNA (fusions) 15 microsatellites HRD status (CE-IVD marked & in a separate panel) |
| Instrument |
NextSeq |
| TAT* |
15 |
| Bone marrow | |
| ctDNA |
X |
| FFPE |
X |
| Genes |
658 |
|---|---|
| Cancer Type |
Solid tumors (important hallmarks of cancer) 638 DNA 20 RNA (fusions) TMB, MSI, HRD |
| Instrument |
NextSeq |
| TAT* |
15 |
| Bone marrow | |
| ctDNA | |
| FFPE |
X |
| Genes | |
|---|---|
| Cancer Type | |
| Instrument | |
| TAT* | |
| Bone marrow | |
| ctDNA | |
| FFPE | |
Taiwan Lab: Our Comprehensive Oncopanels in collaboration with ACT Genomics
| Genes | Cancer Type | Instrument | TAT* | Bone marrow | ctDNA | FFPE | |
|---|---|---|---|---|---|---|---|
|
ACTOnco®+ (DNA-based) |
440 |
Solid tumors (important hallmarks of cancer) TMB |
Ion Torrent |
10 |
X |
||
|
ACTDrug® (DNA-based) |
40 |
Solid tumors (screening of actionable genes) |
Ion Torrent |
10 |
X |
||
|
ACTFusion™ (RNA-based) |
31 |
Solid tumors (actionable fusion genes) |
Ion Torrent |
10 |
X |
||
|
ACTBRCA® (DNA-based) |
2 |
Solid tumors (gene alterations to evaluate PARP inh) |
Ion Torrent |
10 |
X |
X |
|
|
ACTHRD™ (DNA-based) |
24 |
Solid tumors (gene alterations to evaluate PARP inh) |
NextSeq 550 |
10 |
X |
||
|
ACTRisk™ (DNA-based) |
32 |
Identifies genetic alterations related to hereditary cancers |
NextSeq 550 |
10 |
X |
| Genes |
440 |
|---|---|
| Cancer Type |
Solid tumors (important hallmarks of cancer) TMB |
| Instrument |
Ion Torrent |
| TAT* |
10 |
| Bone marrow | |
| ctDNA | |
| FFPE |
X |
| Genes |
40 |
|---|---|
| Cancer Type |
Solid tumors (screening of actionable genes) |
| Instrument |
Ion Torrent |
| TAT* |
10 |
| Bone marrow | |
| ctDNA | |
| FFPE |
X |
| Genes |
31 |
|---|---|
| Cancer Type |
Solid tumors (actionable fusion genes) |
| Instrument |
Ion Torrent |
| TAT* |
10 |
| Bone marrow | |
| ctDNA | |
| FFPE |
X |
| Genes |
2 |
|---|---|
| Cancer Type |
Solid tumors (gene alterations to evaluate PARP inh) |
| Instrument |
Ion Torrent |
| TAT* |
10 |
| Bone marrow | |
| ctDNA |
X |
| FFPE |
X |
| Genes |
24 |
|---|---|
| Cancer Type |
Solid tumors (gene alterations to evaluate PARP inh) |
| Instrument |
NextSeq 550 |
| TAT* |
10 |
| Bone marrow | |
| ctDNA | |
| FFPE |
X |
| Genes |
32 |
|---|---|
| Cancer Type |
Identifies genetic alterations related to hereditary cancers |
| Instrument |
NextSeq 550 |
| TAT* |
10 |
| Bone marrow | |
| ctDNA |
X |
| FFPE | |
Reach out to our genomics team and see how we can help advance your research in the field of Solid Tumor NGS.
Reach out to our genomics team and see how we can help advance your research in the field of Solid Tumor NGS

